Within several ongoing projects, we investigate the structure, function and interactions of proteins and nucleic acids involved in the translation of genetic information. The main emphasis is placed on aminoacyl-tRNA synthetases (aaRS), whose capacity to faithfully translate the standard genetic code and to operate outside of translation are explored.
To reach deep understanding of this essential cellular process in all domains of life, we use multidisciplinary approach which includes enzyme kinetics, biochemical, genetic and biophysical techniques. The extent to which mistranslation occurs is still poorly understood and presents a fundamental question in both basic research and its application to medicine and biotechnology.
We explore aaRS synthetic and proofreading mechanisms against canonical and non-canonical amino acids to unveil molecular foundation of the aminoacylation accuracy and cellular consequences of the disturbed translational fidelity.
The goal is to improve our understanding of the basis for selection of the natural amino acid alphabet, and how violation of the coding principles influences cell physiology. We also aim to engineer translational apparatus for incorporation of synthetic amino acids and production of designer proteins with novel features.
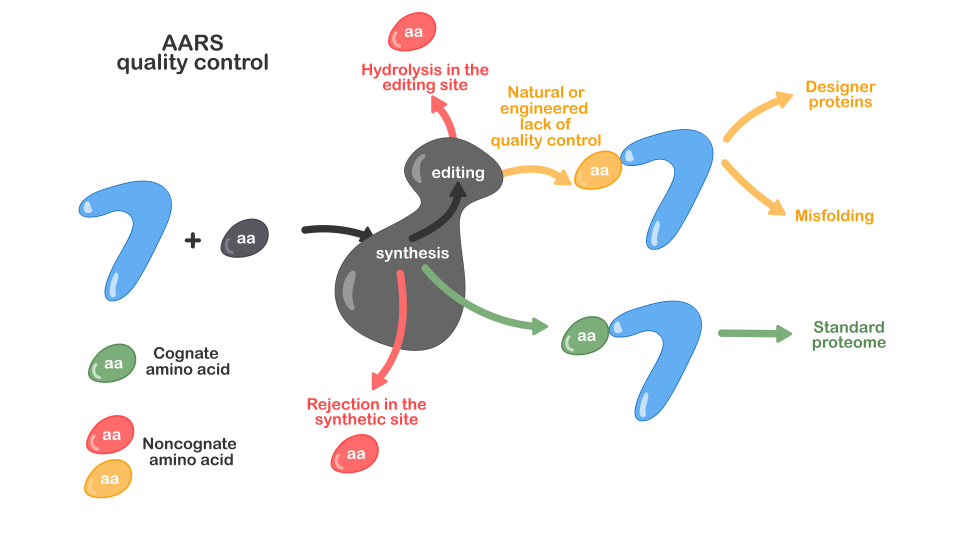
Aminoacyl-tRNA synthetases (AARSs) provide a unique model for studying the evolution of enzyme selectivity. The Achilles heel of AARS selectivity, which is the rejection of non- cognate amino acids at the first encounter, has been addressed by the evolution of editing that clears misrecognized amino acids in the synthetic (pre-transfer editing) and editing (post-transfer editing) active sites. The specificity for a large tRNA substrate, in contrast, relies on the acquisition of a specific tRNA-binding domain. Using biochemical, genetic and enzyme kinetic approaches we aim to understand how AARS selectivity evolved addressing fundamental questions related to the evolution of proteins and biocatalysis. By studying the consequences of AARS errors at the cellular level, we aim to relate requirements for high fidelity with mechanistic solutions evolved to establish the mandatory level of AARS selectivity.
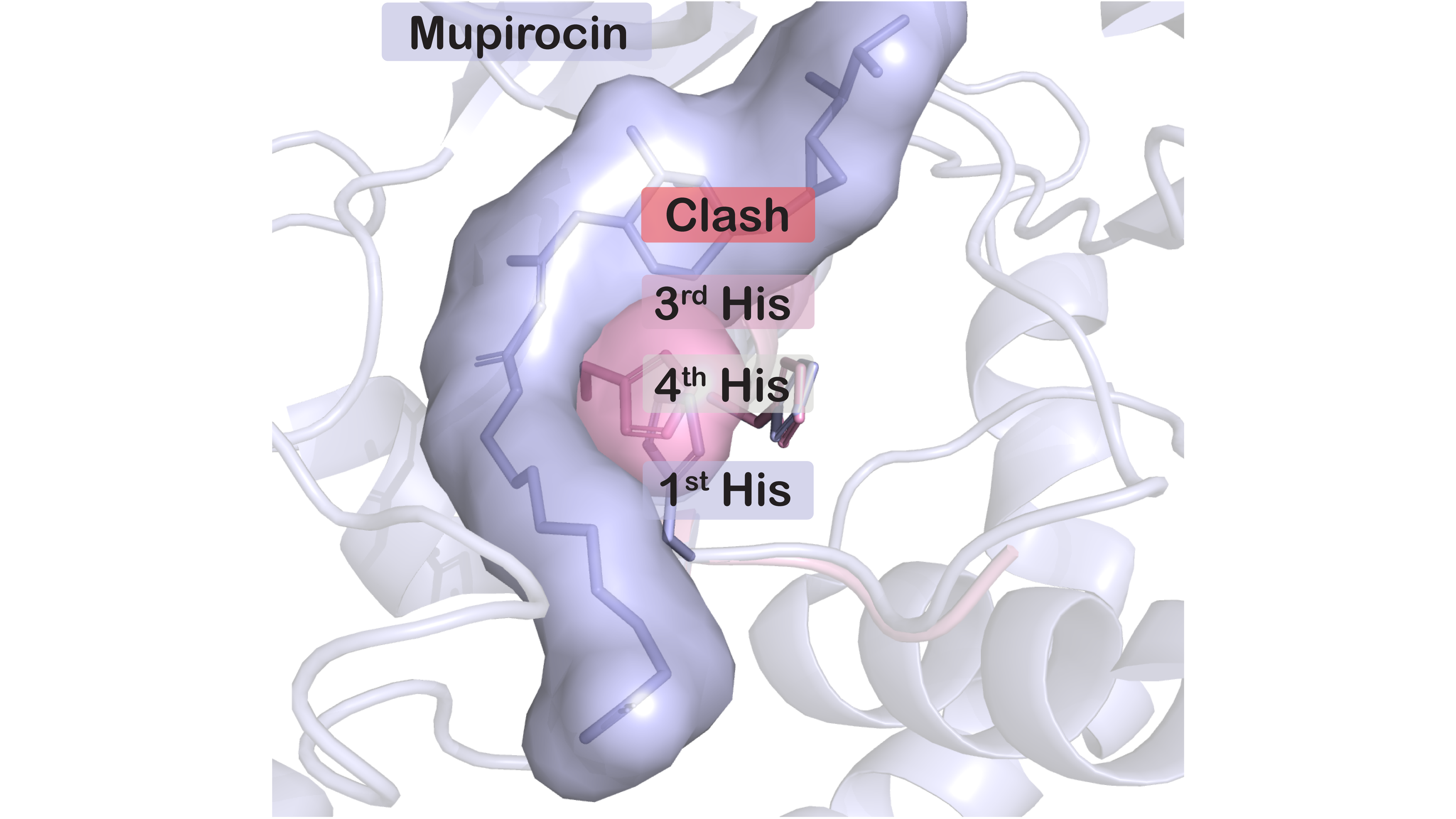
Aminoacyl-tRNA synthetases (AARSs) are fundamental for life and therefore suitable targets for natural and man-made antibiotics. Our recent finding that hyper-resistance to antibiotic mupirocin occurs through natural variation of the most conserved catalytic motif (the HIGH motif) at the IleRS active site without compromising the enzyme’s housekeeping function raises new questions about AARS function and evolution. We aim to explore trade-offs associated with antibiotic (hyper)resistance as well as to identify novel AARS inhibitors among bacterial natural products.